A function to simulate Survival data
sim_adtte.RdThis function simulates survival data with correlated time to progression and overall survival times. Optionally crossover from treatment arms can be simulated.
sim_adtte(
rc_siminfo = FALSE,
rc_origos = FALSE,
id = 1,
seed = 1234,
rho = 0,
pswitch = 0,
proppd = 0,
beta_1a = log(0.7),
beta_1b = log(0.7),
beta_2a = log(0.7),
beta_2b = log(0.7),
beta_pd = log(0.4),
arm_n = 250,
enroll_start = 0,
enroll_end = 1,
admin_censor = 2,
os_gamma = 1.2,
os_lambda = 0.3,
ttp_gamma = 1.5,
ttp_lambda = 2
)Arguments
- rc_siminfo
Should simulation params be included in simulated dataframe (logical). Defaults to FALSE.
- rc_origos
Should OS without switching be included in simulated dataframe (logical). Defaults to FALSE.
- id
Identifer added to simulated dataframe. Defaults to 1.
- seed
Seed used for random number generator. Defaults to 1234.
- rho
correlation coefficient between TTP and OS. Defaults to 0.
- pswitch
proportion of patients who switch. Defaults to 0.
- proppd
proportion of patients with PFS before switch allowed. Defaults to 0.
- beta_1a
treatment effect (as log(Hazard Ratio)) for OS pre PFS. Defaults to log(0.7).
- beta_1b
treatment effect (as log(Hazard Ratio)) for OS post PFS. Defaults to log(0.7).
- beta_2a
treatment effect (as log(Hazard Ratio)) for OS pre PFS (switch). Defaults to log(0.7).
- beta_2b
treatment effect (as log(Hazard Ratio)) for OS post PFS (switch). Defaults to log(0.7).
- beta_pd
treatment effect on progression (as log(HR)). Defaults to log(0.4).
- arm_n
patients per arm. Defaults to 250.
- enroll_start
start of enrollment. Defaults to 0.
- enroll_end
end of enrollment. Defaults to 1.
- admin_censor
end of trial. Defaults to 2.
- os_gamma
weibull shape - for OS. Defaults to 1.2.
- os_lambda
weibull scale - for OS. Defaults to 0.3.
- ttp_gamma
weibull shape - for TTP. Defaults to 1.5.
- ttp_lambda
weibull scale - for TTP. Defaults to 2.
Details
The simulation times are derived from formulas in Austin 2012 adapted to enable correlations between endpoints. Austin, P.C. (2012), Generating survival times to simulate Cox proportional hazards models with time‐varying covariates. Statist. Med., 31: 3946-3958. https://doi.org/10.1002/sim.5452
Examples
require(survival)
require(dplyr)
#> Loading required package: dplyr
#>
#> Attaching package: ‘dplyr’
#> The following objects are masked from ‘package:stats’:
#>
#> filter, lag
#> The following objects are masked from ‘package:base’:
#>
#> intersect, setdiff, setequal, union
ADTTE <- sim_adtte()
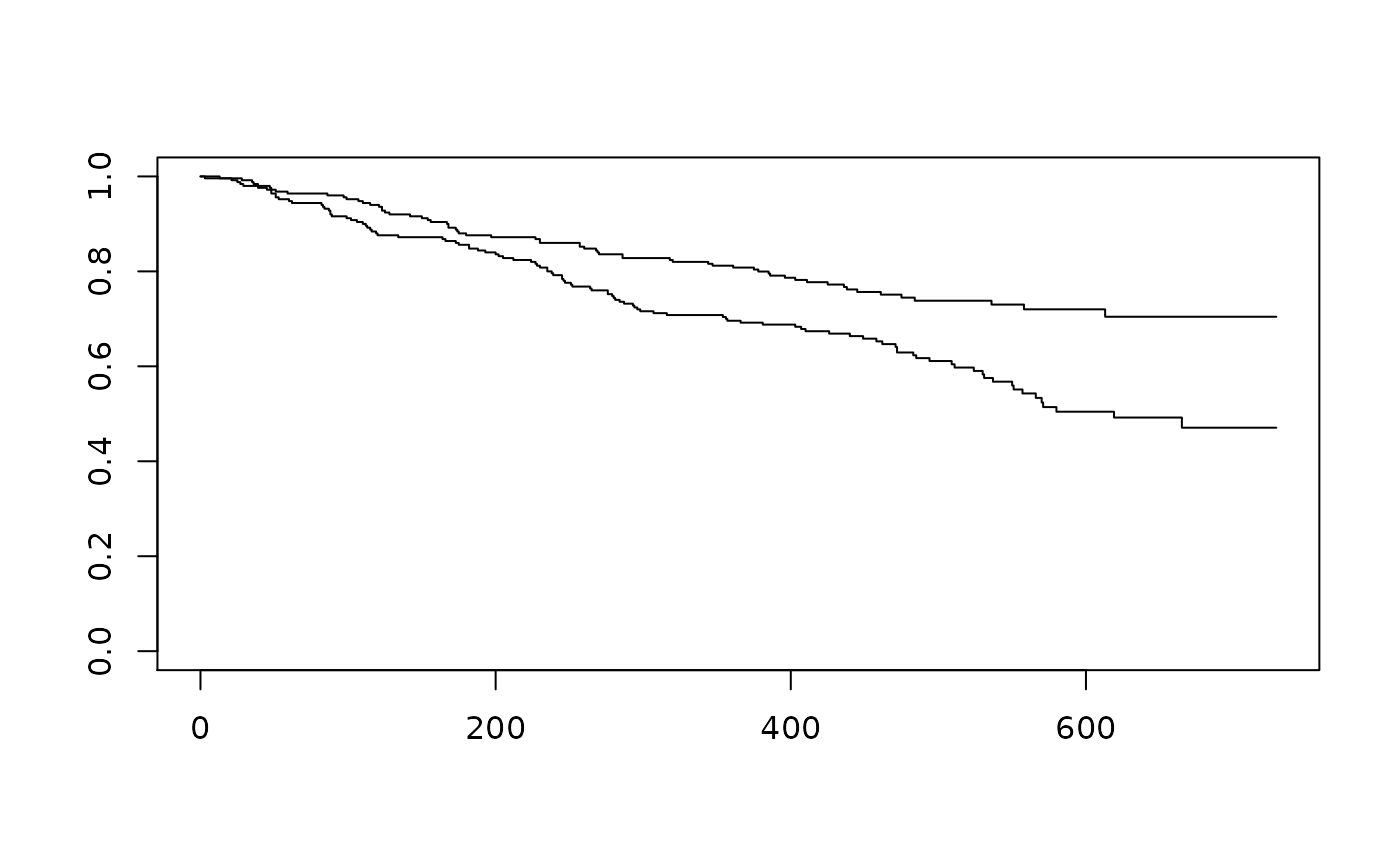
survfit(Surv(AVAL, event = CNSR == 0) ~ ARM, data = filter(ADTTE, PARAMCD == "PFS")) %>%
plot()
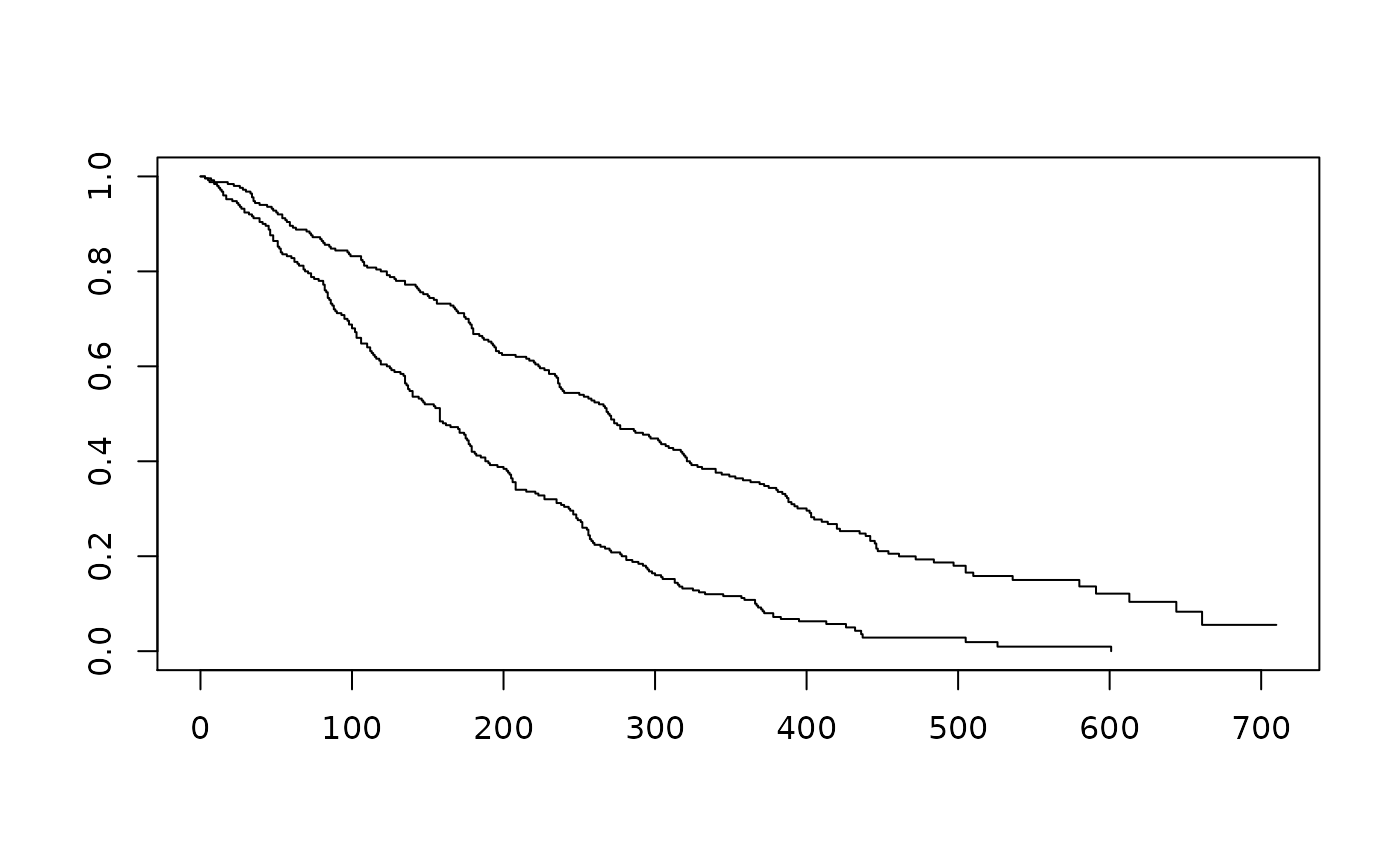 survfit(Surv(AVAL, event = CNSR == 0) ~ ARM, data = filter(ADTTE, PARAMCD == "OS")) %>%
plot()
survfit(Surv(AVAL, event = CNSR == 0) ~ ARM, data = filter(ADTTE, PARAMCD == "OS")) %>%
plot()